
© Columbia University, all rights reserved.

Quantitative Proteomics and Metabolomics Center

Mascot proteomics software (V. 2.8.1) (Matrix Science Ltd.)
Loaded on a Lenovo ThinkStation P500 with Intel Xeon E5-1650 v3 Processor (15MB Cache, 3.50GHz), 64 GB RAM. This database server allows comparison between mass spectra and protein databases to facilitate the identification of proteins.
Loaded on a Lenovo ThinkStation P500 with Intel Xeon E5-1650 v3 Processor (15MB Cache, 3.50GHz), 64 GB RAM. This database server allows comparison between mass spectra and protein databases to facilitate the identification of proteins.
Orbitrap Exploris 240 mass spectrometer (Thermo Scientific)
This is a high performance benchtop quadrupole Orbitrap system that combines a quadrupole, an ion routing multipole, and Orbitrap analyzer and features HCD and in-source fragmentation. The high-field Orbitrap Mass Analyzer with enhanced FT transient processing resulting in resolving power from 15,000 up to 240,000 FWHM at m/z 200 and isotopic fidelity, m/z range of 40-6,000 with up to 22 Hz acquisition speed.
This is a high performance benchtop quadrupole Orbitrap system that combines a quadrupole, an ion routing multipole, and Orbitrap analyzer and features HCD and in-source fragmentation. The high-field Orbitrap Mass Analyzer with enhanced FT transient processing resulting in resolving power from 15,000 up to 240,000 FWHM at m/z 200 and isotopic fidelity, m/z range of 40-6,000 with up to 22 Hz acquisition speed.
Major Equipment and Software Resources in the Center
Proteome Discoverer Software V. 1.4 (Thermo Scientific)
-For Thermo Orbitrap Q Exactive HF. Supports multiple database search algorithms including SEQUEST and Mascot for comprehensive analyses. This software runs on our HP ProLiant ML350p Windows server with two Intel Xeon E5-2650v2 (2.6GHz/8-core) processors with 128 GB RAM and dual 600 GB SAS 15K rpm enterprise drives.
-For Thermo Orbitrap Q Exactive HF. Supports multiple database search algorithms including SEQUEST and Mascot for comprehensive analyses. This software runs on our HP ProLiant ML350p Windows server with two Intel Xeon E5-2650v2 (2.6GHz/8-core) processors with 128 GB RAM and dual 600 GB SAS 15K rpm enterprise drives.


PEAKS Studio
PEAKS Studio 11.5 build 20230821 (Bioinformatics Solutions, Inc.). PEAKS studio runs on our in-house Windows server on a Dell Precision 7860 server with Dual: Intel Xeon Platinum 8480+, 56 cores,, (total 112 cores), 192 GB RAM and NAS array with 32 TB (8 x 4 Tb Drives). In parallel we run PEAKS concurrently supporting analysis of additional data on a Dell Precision 7820 with dual Intel Xeon Gold 6246R 3.4GHz,16C, (total 32 cores),192 GB RAM, M.2 1TB PCIe NVMe solid state Drive and 10 TB of SATA storage. The PEAKS systems performs data management and analysis for label-free and labeled mass spectrometry data for accurate peptide and protein identifications and quantitation.
Data mining matches accurate mass and retention time measurements for mass spectrometric features across large and complex experiments, substantially increasing usable data from each experiment. In addition, Peaks Studio provides statistical analysis and visualization tools for protein expression analysis. PEAKS studio provides advanced capabilities including de novo sequencing for identification of sequences not present in a database, spectral library search to enhance the effectiveness of data-independent acquisition, post-translational modification (PTM) searches, and sequence variant and mutation searches with the SPIDER algorithm. software is fully enabled to import both raw and data, fully supporting our data-independent acquisition and data-dependent acquisition workflows for the Orbitrap Exploris 240 and Orbitrap Q Exactive HF
PEAKS Studio 11.5 build 20230821 (Bioinformatics Solutions, Inc.). PEAKS studio runs on our in-house Windows server on a Dell Precision 7860 server with Dual: Intel Xeon Platinum 8480+, 56 cores,, (total 112 cores), 192 GB RAM and NAS array with 32 TB (8 x 4 Tb Drives). In parallel we run PEAKS concurrently supporting analysis of additional data on a Dell Precision 7820 with dual Intel Xeon Gold 6246R 3.4GHz,16C, (total 32 cores),192 GB RAM, M.2 1TB PCIe NVMe solid state Drive and 10 TB of SATA storage. The PEAKS systems performs data management and analysis for label-free and labeled mass spectrometry data for accurate peptide and protein identifications and quantitation.
Data mining matches accurate mass and retention time measurements for mass spectrometric features across large and complex experiments, substantially increasing usable data from each experiment. In addition, Peaks Studio provides statistical analysis and visualization tools for protein expression analysis. PEAKS studio provides advanced capabilities including de novo sequencing for identification of sequences not present in a database, spectral library search to enhance the effectiveness of data-independent acquisition, post-translational modification (PTM) searches, and sequence variant and mutation searches with the SPIDER algorithm. software is fully enabled to import both raw and data, fully supporting our data-independent acquisition and data-dependent acquisition workflows for the Orbitrap Exploris 240 and Orbitrap Q Exactive HF
Orbitrap Q Exactive HF mass spectrometer (Thermo Scientific)
This leading edge instrument has a resolution of 240,000 and 18 Hz scan speed. This system allows extraordinarily deep proteome searches as well as intensive studies of post-translational modifications
This leading edge instrument has a resolution of 240,000 and 18 Hz scan speed. This system allows extraordinarily deep proteome searches as well as intensive studies of post-translational modifications



Our Campus
ACQUITY I-Class UltraPerformance Liquid Chromatograph (UPLC) with Sample Manager (Waters Corp.)
ACQUITY I-Class UltraPerformance Liquid Chromatograph (UPLC) with Sample Manager (Waters Corp.) The ACQUITY UPLC I-Class features flexibility and precise sampling capabilities. It provides high-precision injections, excellent sample recovery, and optimal performance. This system supports fractionation of complex peptide mixtures with the attached BioFrac Fraction Collector (Bio-Rad Laboratories).
ACQUITY I-Class UltraPerformance Liquid Chromatograph (UPLC) with Sample Manager (Waters Corp.) The ACQUITY UPLC I-Class features flexibility and precise sampling capabilities. It provides high-precision injections, excellent sample recovery, and optimal performance. This system supports fractionation of complex peptide mixtures with the attached BioFrac Fraction Collector (Bio-Rad Laboratories).
Our Lab




Vanquish Neo UHPLC
Coupled to the Exploris 240 is a Vanquish Neo UHPLC system (Thermo Scientific) is the all-in-one nano-, capillary-, and micro-flow LC for high-sensitivity LCMS workflows supporting direct and trap-and-elute injections for low flow applications from 0.001-100 µL/min at max. 1500 bar.
Coupled to the Exploris 240 is a Vanquish Neo UHPLC system (Thermo Scientific) is the all-in-one nano-, capillary-, and micro-flow LC for high-sensitivity LCMS workflows supporting direct and trap-and-elute injections for low flow applications from 0.001-100 µL/min at max. 1500 bar.
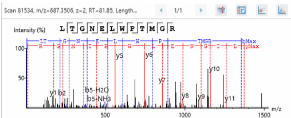
Supporting the Q Evactive HF mass spectrometer is an RSLCnano UPLC also
from Thermo Scientific equipped with an autosampler, column oven and
50-cm analytical column for exceptional chromatographic separations
